Abstract
Introduction. Minimal residual disease (MRD) is the most powerful prognostic factor in acute lymphoblastic leukemia (ALL). Currently, real-time quantitative PCR (RQ-PCR) is the most widely used molecular method for MRD assessment, rigorously standardized within the EuroMRD consortium. According to the EuroMRD guidelines (Van der Velden et al. Leukemia 2007), a non-negligible fraction of patients with very low MRD levels are classified as positive not-quantifiable (PNQ), a definition that may result problematic in the clinical practice. Digital-droplet-PCR (ddPCR) allows an absolute quantification without the need of a standard curve and has the potential to overcome some limitations of RQ-PCR. High degrees of efficiency, sensitivity and accuracy have been reported for ddPCR compared to RQ-PCR, but no established guidelines for ddPCR MRD analysis and interpretation have so far been defined and its ability to correctly evaluate very low MRD levels is still under investigation.
In the present study, we assessed MRD by ddPCR in pediatric ALL cases classified as PNQ and/or negative by RQ-PCR at days +33 and/or +78 of the AIEOP-BFM ALL 2000 trial, to evaluate the potential of ddPCR for low MRD quantification and patients' risk stratification.
Patients and Methods. A total of 211 pediatric ALL patients enrolled in the AIEOP-BFM ALL 2000 trial were included in the study. We analyzed 124 B-lineage ALL patients defined as intermediate risk (IR) who had high positive MRD at day +33 and at day +78 were either PNQ (n=45, Slow Early Responders (SER)) or negative (n=79). A case-control design was applied to 36 B- and T-lineage relapsed ALL patients (cases) who at day +33 had PNQ MRD (n=12, IR) or were negative (n=24, standard risk (SR)) and to matched controls (21 and 30 patients who did not present a relapse). ddPCR analysis was performed as previously published (Della Starza et al, BJH 174, 541-9, 2016), by using 1.5 μg and 3.0 μg DNA of the follow-up samples. In the absence of an international consensus, data have been analyzed using two alternative guidelines; results are reported according to Della Starza et al (BJH 2016).
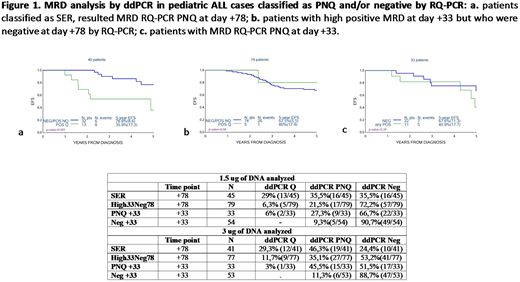
Results. Among 45 SER patients, ddPCR performed on 1.5 μg DNA of PNQ samples at day +78 revealed that 13 were quantifiable (Q), 16 PNQ and 16 negative (NEG) . When 3.0 μg of DNA were used (41/45 samples due to material availability), 12 were Q, 19 PNQ and 10 NEG. Event-free survival (EFS) curves are shown in Fig. 1a.
Among the 79 patients with high positive MRD at day +33 but who were negative at day +78, ddPCR on 1.5 μg DNA of day +78 identified 5 as Q, 17 PNQ and 57 NEG. When 3.0 μg DNA was used (77/79 samples), 9 patients were Q, 27 PNQ and 41 NEG. EFS curves are reported in Fig. 1b.
When ddPCR was applied to 33 PNQ samples at day +33, 2 were Q, 9 PNQ and 22 NEG; when using 3.0 μg of DNA, 1 was Q, 15 were PNQ and 17 NEG. EFS curves are shown in Fig. 1c.
Lastly, ddPCR on 1.5 μg of day +33 DNA of 54 SR patients showed 5 PNQ and 49 NEG, whilst by using 3.0 μg on 53 sample, 7 were PNQ and 46 NEG.
Conclusions. Our data demonstrate that ddPCR is a very promising tool for the evaluation of MRD in ALL cases with very low or negative RQ-PCR MRD results. In particular, among SER patients most relapses occurred in cases with quantifiable ddPCR MRD at day +78, while patients with negative or PNQ MRD by ddPCR at day +78 had a better outcome. Based on these results, high-risk treatment could be offered only to ddPCR quantifiable cases. Among patients with highly positive MRD at day +33 and negative at day +78, the small number of cases with quantifiable disease by ddPCR at present does not allow to establish the impact of quantification; consistently with SER patients, the outcome was similar for patients with negative or PNQ MRD by ddPCR at day +78. Similarly, among patients with PNQ MRD by RQ-PCR at day +33, a similar outcome was observed for cases negative or PNQ by ddPCR. Lastly, in most SR patients ddPCR confirmed the negative results of RQ-PCR at day +33, associated with an extremely good kinetics of disease reduction, independently of the MRD PCR method. Overall, our data indicate that ddPCR is as sensitive as RQ-PCR and can provide a potentially more accurate prognostic stratification for cases defined as PNQ MRD by RQ-PCR, in view of its ability to quantify without a standard curve. The application of ddPCR in a prospective clinical protocol with international guidelines is needed to define whether it can result in an overall improvement of pediatric ALL patients' stratification and outcome.
Foà:AMGEN: Other: ADVISORY BOARD; JANSSEN: Other: ADVISORY BOARD, Speakers Bureau; CELTRION: Other: ADVISORY BOARD; INCYTE: Other: ADVISORY BOARD; ABBVIE: Other: ADVISORY BOARD, Speakers Bureau; ROCHE: Other: ADVISORY BOARD, Speakers Bureau; NOVARTIS: Speakers Bureau; CELGENE: Other: ADVISORY BOARD, Speakers Bureau; GILEAD: Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal